We've published a new paper
We’re proud to announce that Konsta and Divyesh’s manuscript on transposable elements as cancer-specific enhancers was published in Nature Communications on 1st September, 2023. Huge congrats to all co-authors and collaborators!
Read more about our research findings below.
Mobile DNA elements can boost the development of malignant tumours
New research findings pertaining to transposable elements, also known as jumping genes or mobile elements, increase our understanding of how cancers develop.
Nearly half of the human genome is composed of transposable elements, which are sequences of DNA that are able to replicate and relocate in the genome. Previously, transposable elements have been considered ‘junk’ DNA in the human genome.
However, research has shown that the ability to replicate and move has enabled these sequences to develop into new regulatory regions during evolution, particularly in early stages of development. In our study, we investigated how cancer cells can hijack these mobile elements to boost tumours. The findings revealed new information on the development of cancers.
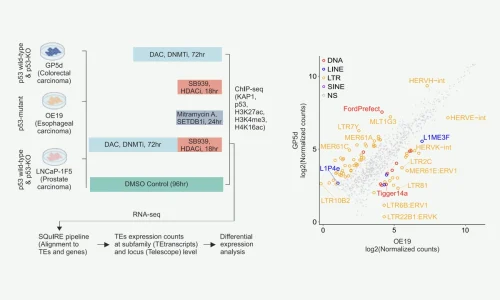
We identified specific families of transposable elements that are active in liver and colon cancers. These elements have binding sites for specific transcription factors, which are proteins that control gene expression through the regulatory elements.
We demonstrated that these specific transposable elements boost gene regulation in a cancer-specific manner. In other words, different cancer types take over, as it were, the elements for their own regulatory purposes.Konsta Karttunen
New knowledge on the regulatory roles of transposable elements in cancer
Prior research has already shown that transposable elements play a significant role in gene regulation, for example, during human embryonic development.
However, because of their replication in the genome, these elements are difficult to study, since the genome contains several copies of nearly identical elements. Cells also actively strive to keep transposable elements silent and non-functional.
We employed a method called massively parallel reporter assay that can be used to directly identify non-coding regulatory regions in the human genome. Previously, this technique has only rarely been used to investigate the regulatory roles of transposable elements, particularly in the case of cancers.
Our research sheds light on regulatory roles more broadly than previously thought.Divyesh Patel
Mobile DNA elements can be utilised in early cancer diagnostics
A better understanding of mobile elements can help in developing therapies for various cancers.
The transposable elements activated in cancers could be used as markers that help in diagnosing cancer in its early stages. Moreover, the cellular immune reaction brought about by transposable elements can be utilised in immunotherapies for cancer.
Biswajyoti Sahu
Publication Abstract
Transposable elements (TE) are repetitive genomic elements that harbor binding sites for human transcription factors (TF). A regulatory role for TEs has been suggested in embryonal development and diseases such as cancer but systematic investigation of their functions has been limited by their widespread silencing in the genome.
Here, we utilize unbiased massively parallel reporter assay data using a whole human genome library to identify TEs with functional enhancer activity in two human cancer types of endodermal lineage, colorectal and liver cancers.
We show that the identified TE enhancers are characterized by genomic features associated with active enhancers, such as epigenetic marks and TF binding.
Importantly, we identify distinct TE subfamilies that function as tissue-specific enhancers, namely MER11- and LTR12-elements in colon and liver cancers, respectively. These elements are bound by distinct TFs in each cell type, and they have predicted associations to differentially expressed genes.
In conclusion, these data demonstrate how different cancer types can utilize distinct TEs as tissue-specific enhancers, paving the way for comprehensive understanding of the role of TEs as bona fide enhancers in the cancer genomes.
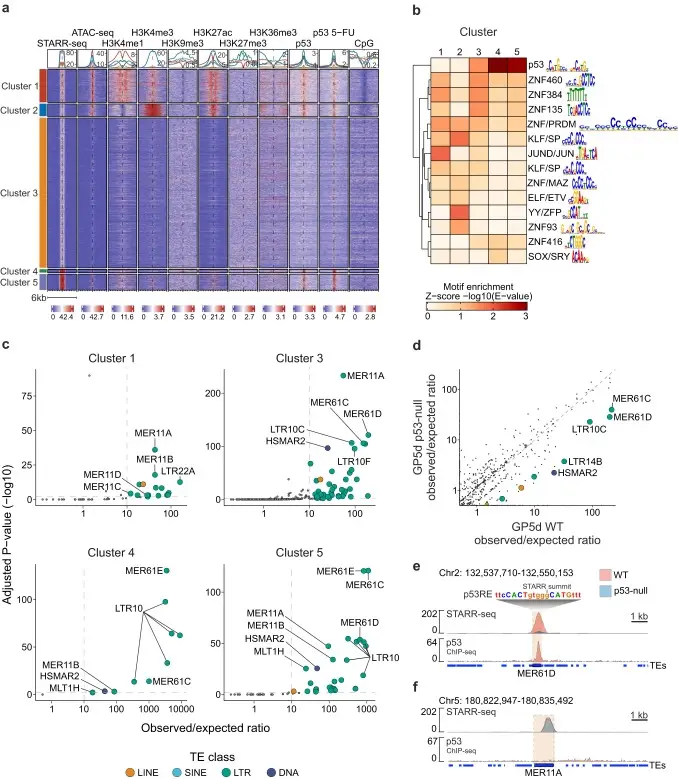
differentially enriched TE subfamilies.